Rodent Genome and Pathogen Atlas
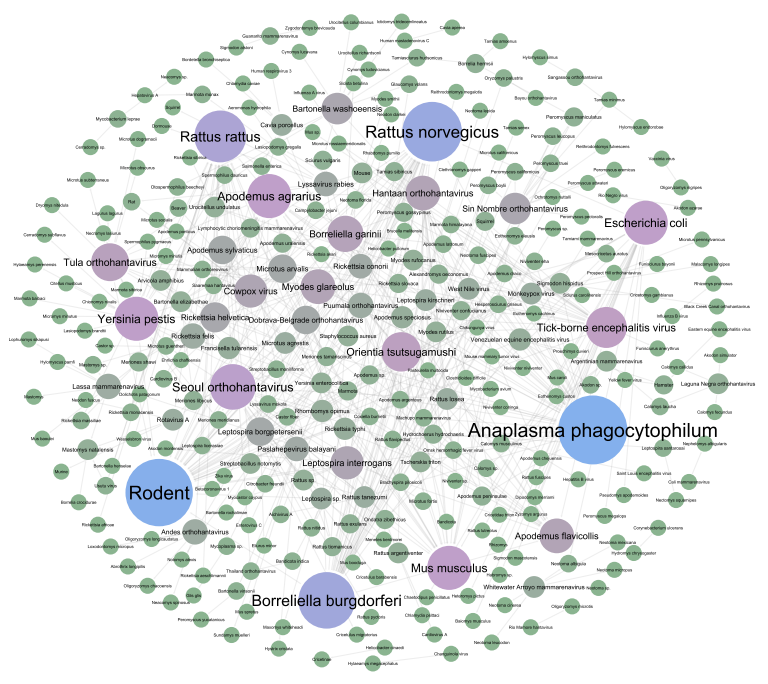
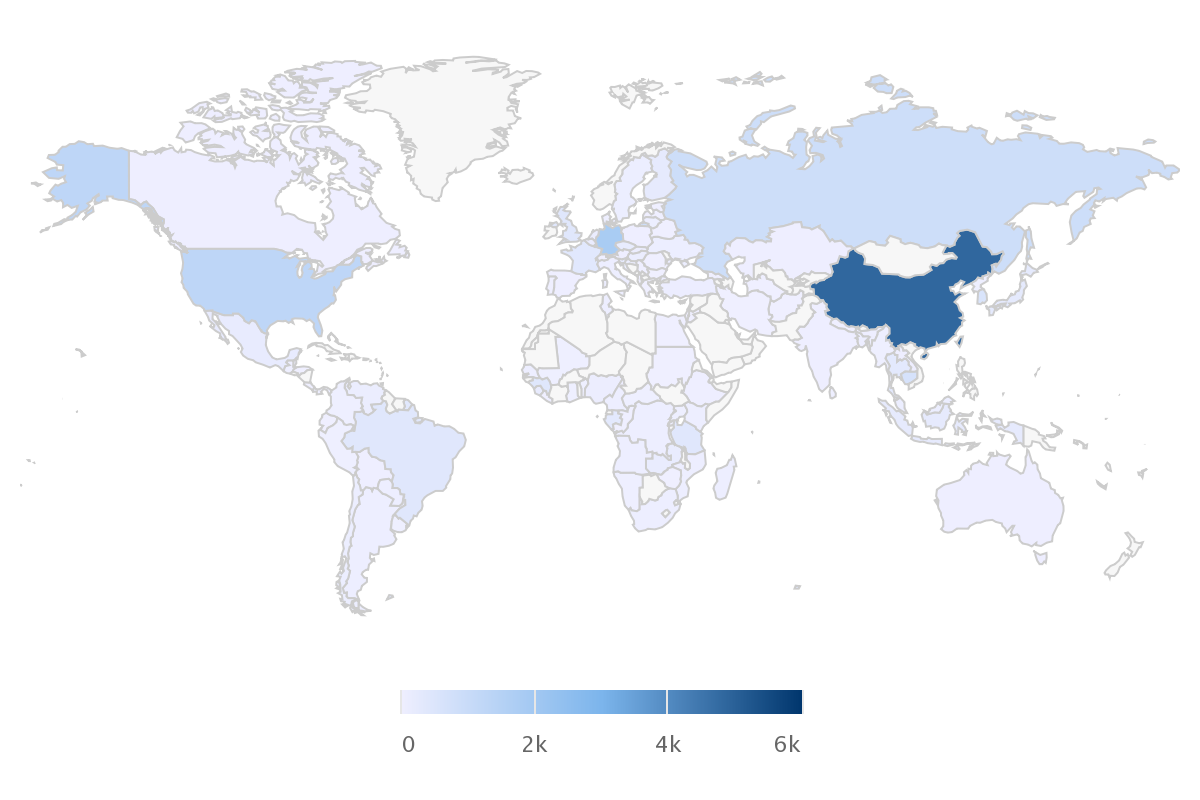
The Rodent Genome and Pathogen Atlas is a multi-omics tool with the aim of facilitating insights into human medical research. The RGPdb systematically collects genomic and pathogens resources of rodents. By integrating diverse omics data and developing sophisticated analysis tools, it has established the most comprehensive and functionally rich rodent omics analysis platform to date.
Navigation

Species
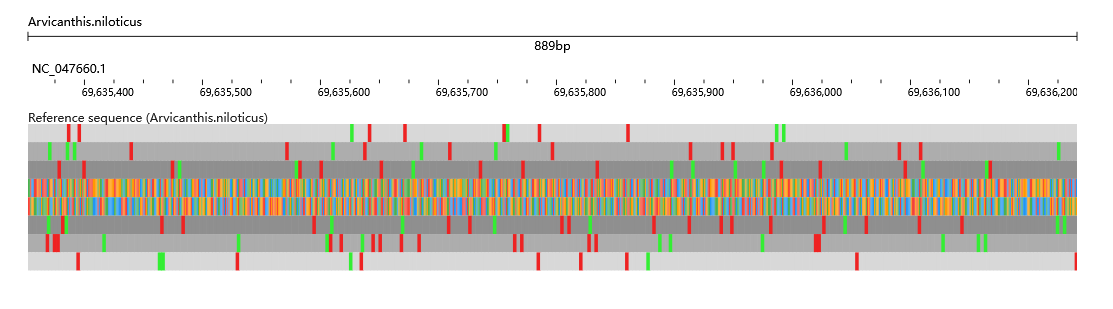
This module is a tool designed for analyzing and interpreting species data.
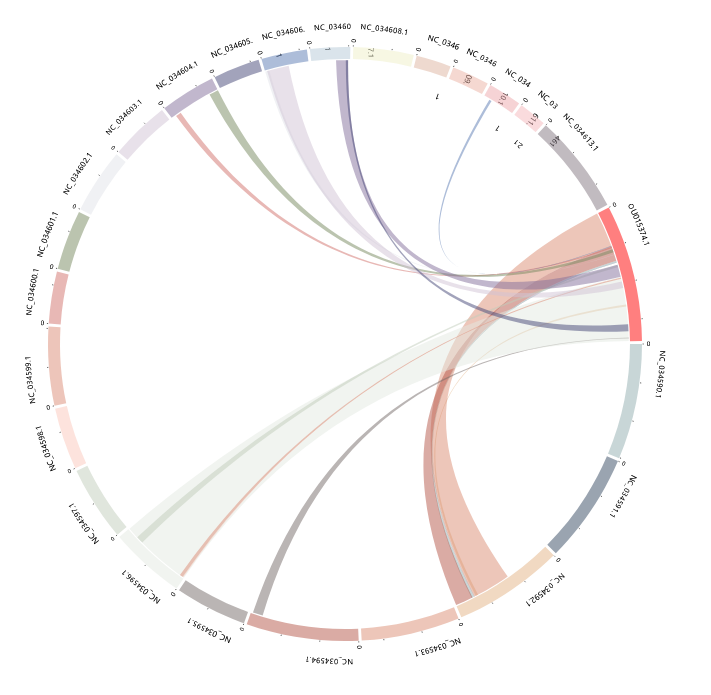
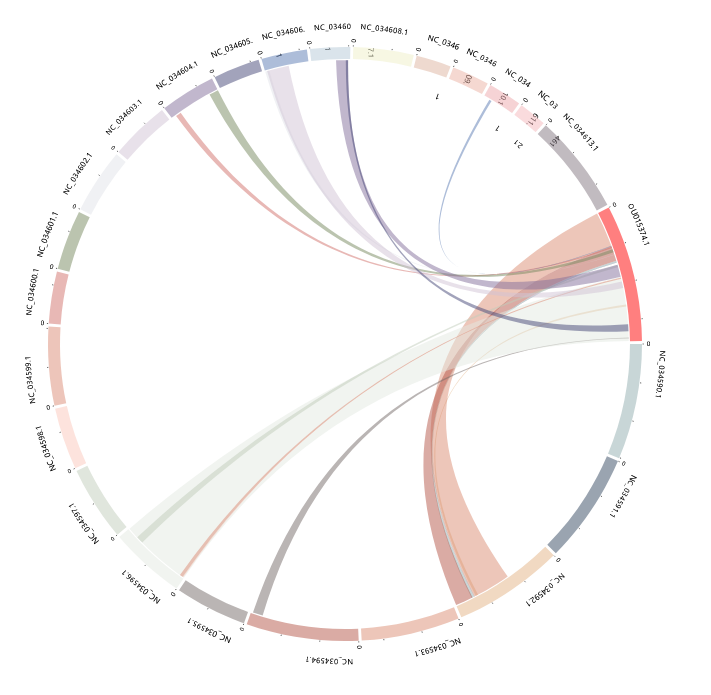
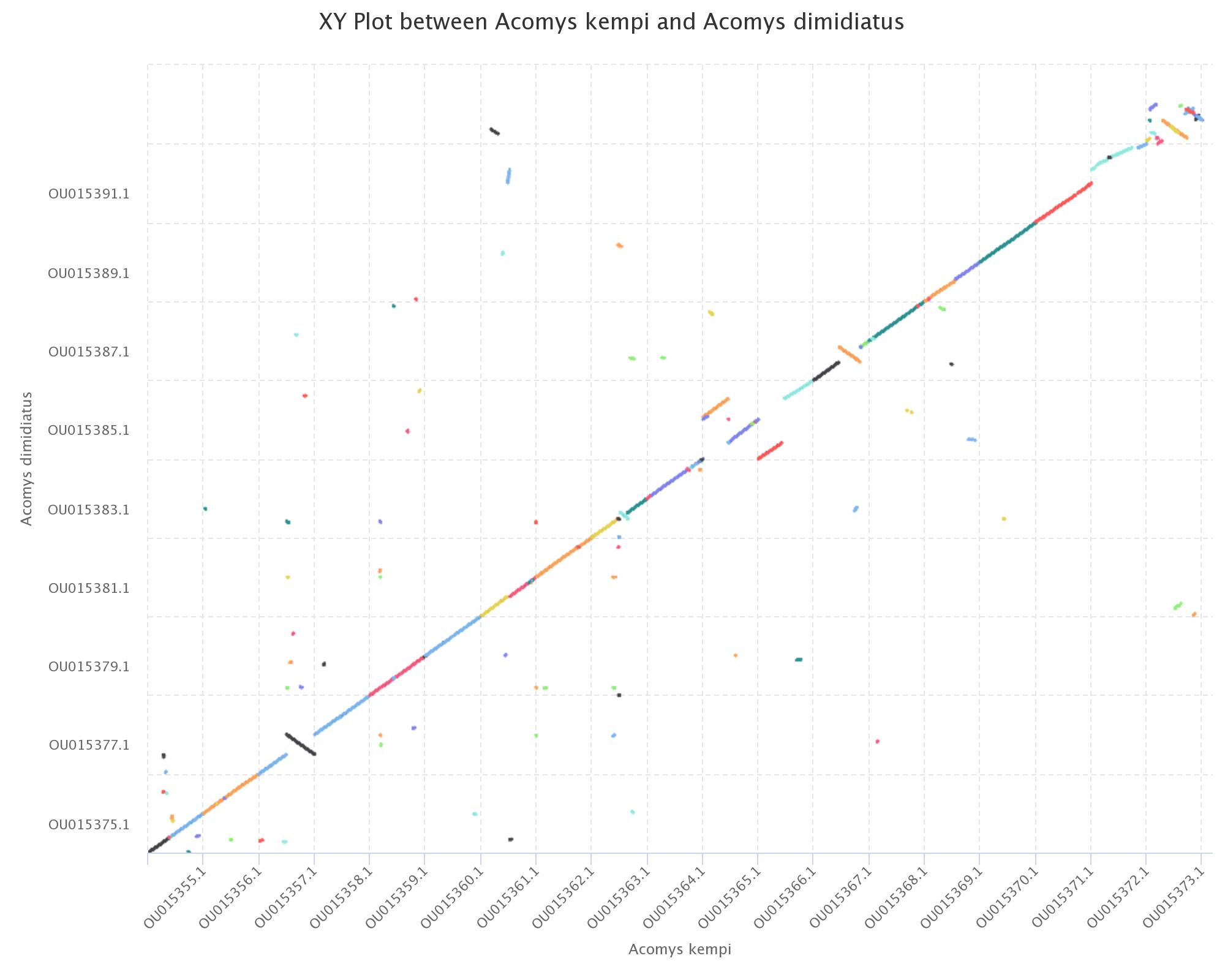
Synteny
This module is a tool designed for analyzing and interpreting synteny data.
Search
This module is a tool designed for search and seq fetch species‘ gene info.

BLAST
BLAST (basic local alignment search tool) is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA and/or RNA sequences.

Download
This module is a tool designed for download species data(Genome,CDS,Protein,GFF,Annotion).
Document
This module is a tool designed to view database information and our team information.
Tools

Muscle
MUSCLE is one of the best-performing multiple alignment programs according to published benchmark tests, with accuracy and speed that are consistently better than CLUSTALW.

Genewise
GeneWise compares a protein sequence to a genomic DNA sequence, allowing for introns and frameshifting errors.

Primer
Primer3 is a widely used program for designing PCR primers (PCR = "Polymerase Chain Reaction"). PCR is an essential and ubiquitous tool in genetics and molecular biology.
Lastz
This document describes installation and usage of the LASTZ sequence alignment program. LASTZ is a drop-in replacement for BLASTZ, and is backward compatible with BLASTZ's command-line syntax.

GO Enrichment
GO enrichment analysis is to classify differential genes according to GO, and conduct significance analysis, misjudgment rate analysis and enrichment analysis based on discrete distribution on the classification results.

KEGG Enrichment
KEGG Enrichment Analysis is often used to study gene function and screen key genes in differential gene sets.